NovaSeq 6000Dx: TSO 500 Auto-launch Analysis in Cloud
This section provides a guided example of the end-to-end user flow for a sequencing run for TruSight Oncology 500 on NovaSeq 6000Dx (RUO mode). The workflow includes automated data streaming to cloud and auto-launch of secondary analysis minimizing manual touchpoints. The tutorial covers:
Instrument and BaseSpace configuration
Run Set-up, start and monitoring
Accessing data in ICA and BaseSpace
Instrument and BaseSpace Configuration
Configure BaseSpace
Follow BaseSpace Setup instructions to configure sequencing run upload to ICA.
Configure BaseSpace connection on NovaSeq 6000Dx
On the RUO side of the instrument, select Instrument Settings.

Select RUO SETTINGS tab to configure BaseSpace connection.
Select the box BaseSpace Sequence Hub: (Optional to upload and store run data in cloud server)
Select Run Monitoring and Storage (image below) in the hosting location dropdown menu
Choose the region where BaseSpace is hosted from the hosting location dropdown menu

Specify the domain name. It is only possible to use a private domain. Contact the IT department to add the correct URLs to the firewall allow list. For further information, please refer to Network Connections and Control Computer Firewall.

Run Set-up, start and monitoring
Plan the Sequencing Run
BaseSpace Run Planning provides an interface for planning a sequencing run.
Navigate to BaseSpace and change to your desired workgroup or personal context. This requires you to login to your domain first.
Navigate to the "Runs" tab in BaseSpace. Select the "New Run" button and select "Run Planning".
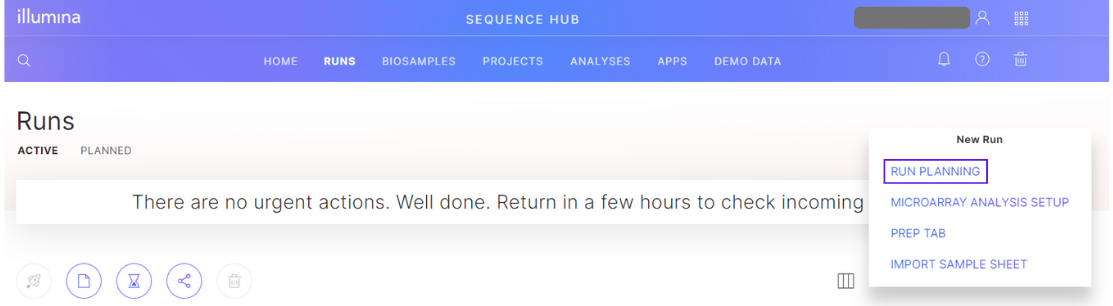
Create a run
Use the BaseSpace Run Planning tool to create a planned run.
Set the Analysis Location to "BaseSpace/Illumina Connected Analytics" to configure for Cloud analysis.

The configuration input forms vary depending on the instrument, secondary analysis pipeline, and other selections (see Sequencer Auto-launch Analyses Compatibility for details). The images below demonstrate screens using a NovaSeq 6000 Dx instrument with DRAGEN TSO500 analysis.

Select "DRAGEN TSO 500 Analysis Software - 2.5.2" or "DRAGEN TruSight Oncology 500 Analysis Software - 2.5.2 (with HRD)" from the Application drop-down menu.
Select TruSight Oncology 500 or TruSight Oncology 500 High Throuput From the Library Prep Kit drop-down menu.
Click Next to navigate to the next step.

Enter the names of the samples in the run. Alternatively, you can download a sample template by clicking "DOWNLOAD TEMPLATE" as indicated in the image below, then use it to import the sample information.

In the Per Sample Configuration section, add Sample Type, Pair ID, and Sample Feature. For HRD sample, specify "HRD" in the Sample Feature field, otherwise leave it empty.

Upon completion, export the sequencing run as a SampleSheet.csv file.
Start the Sequencing Run
Follow the instructions for the sequencer to execute the planned sequencing run for cloud analysis. Attach the SampleSheet.csv saved from the previous step.

Monitor the Sequencing Run
After a sequencing run is started, use BaseSpace to monitor the run through completion. Use the BaseSpace Run Summary view to track the run progress.
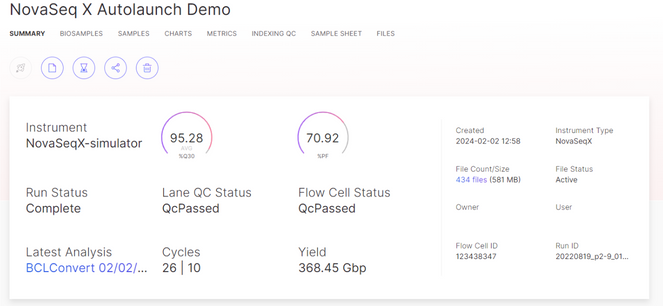
View Sequencing Run Output
Refer to Sequencer Run Data in ICA for guidance on viewing the sequencing run data in ICA.
View Secondary Analysis Results
When secondary analysis finishes, the results can be viewed in BaseSpace in the corresponding project. The result files can be viewed and downloaded from the Files tab. In addition, the files can be downloaded using BaseSpace Sequence Hub CLI (BaseSpace CLI) tool.
Last updated
Was this helpful?
